m6A methylation in the perinatal field
Kosuke Taniguchi, Tomoko Kawai
DOI10.36648/2471-805X.7.1.62
Kosuke Taniguchi and Tomoko Kawai*
Department of Maternal-Fetal Biology, National Research Institute for Child Health and Development, 2-10-1, Okura, Setagaya-ku, Tokyo, 157-8535 Japan.
- *Corresponding Author:
- Tomoko Kawai
Department of Maternal-Fetal Biology,
National Research Institute for Child Health and Development, 2-10-1,
Okura, Setagaya-ku, Tokyo, 157-8535 Japan,
Tel: +81-3-3416-0181;
E-mail: kawai-tm@ncchd.go.jp
Received Date: December 04, 2020;Accepted Date: December 18, 2020;Published Date: December 25, 2020
Citation: Kawai T, Taniguchi K (2020) m6A Methylation in the Perinatal Field. Pediatric Care Vol.7 No.1: 2
N6-Methyladenosine Overview
N6-methyladenosine (m6A), a major modification of messenger RNA (mRNA) and long non-coding RNA, plays critical roles in RNA metabolism and function [1]. Among the many types of RNA modification, m6A is the most frequent and abundant chemical post-transcriptional RNA modification [2]. Methylated RNA immunoprecipitation followed by sequencing (MeRIP-Seq) is a comprehensive assay to determine the presence of m6A [1]. Since the development of this assay, the functions of m6A have gradually become clearer m6A within coding mRNA are most abundantly observed in the vicinity of the stop codon, especially within the 3'UTR, and they have a consensus sequence of RRACH, where R is a purine and H is any base except for G [1]. m6A is involved in post-transcriptional regulation, especially in determining the stability and lifespan of mRNA [3]. Furthermore, several m6A regulators have been reported, such as the methylating enzyme m6A writer proteins (METTL3, METTL14, METTL16, WTAP), the demethylating enzyme m6A eraser proteins (FTO, ALKBH5), and m6A reader proteins (YTH family) that recognize m6A [3]. The m6A site in mRNA of the same gene differs between cell and tissue types; m6A levels change in response to external stimuli, thereby functioning as a dynamic type of modification that fine-tunes gene expression [4]. Notably, although the levels of mRNA (arising from gene expression) and protein are positively correlated, the correlation is weak; it is not a perfect correlation [5]. Therefore, elucidation of post-transcriptional mRNA regulation through m6A modifications can help clarify the role of genes involved in various cellular events.
Abbreviations
AFD: Appropriate For Date; E: Embryonic Day; m6A: N6- methyladenosine; mRNA: messenger RNA; MeRIP-Seq: Methylated RNA Immunoprecipitation followed by Sequencing; PE: Preeclampsia; SNPs: Single Nucleotide Polymorphisms; SFD: Small For Date.
Development and m6A in Reproductive Stage
So far, several cellular events that are essential for life have been suggested to be under the regulation of m6A modification. For example, Mettl3-null mutant mouse embryos died by embryonic day (E) 6.5, whereas E3.5 knockout blastocysts retained normal characteristics [6]. Depletion of Mettl14, another component of the m6A writer complex, could only develop embryos up to E6.5 [7]. Similarly, knockout of newly identified Mettl16 in mice allows embryonic development until the blastocyst stage, but causes development arrest around the time of implantation [8]. These studies suggest that m6A methyltransferases are indispensable for early development. However, the m6A reader YTHDF2 is intrinsically required for female fertility. Oocytespecific deletion of Ythdf2 produced normal numbers of MII oocytes, and was fertilized. However, development was derailed at or prior to the two-cell stage in zygotes derived from Ythdf2- deleted oocytes. YTHDF2 is required to instruct the appropriate transcript dosage during oocyte maturation [9], which suggests that m6A modification and recognition are essential within this process as well. Moreover, it was revealed that mice deficient in Alkbh5 have compromised spermatogenesis, which likely results in apoptosis of pachytene and metaphase-stage spermatocytes [10]. This suggests that m6A demethylation is required for spermatogenesis. These studies all suggested essential roles of m6A in the reproductive stage.
Human Placenta and m6A
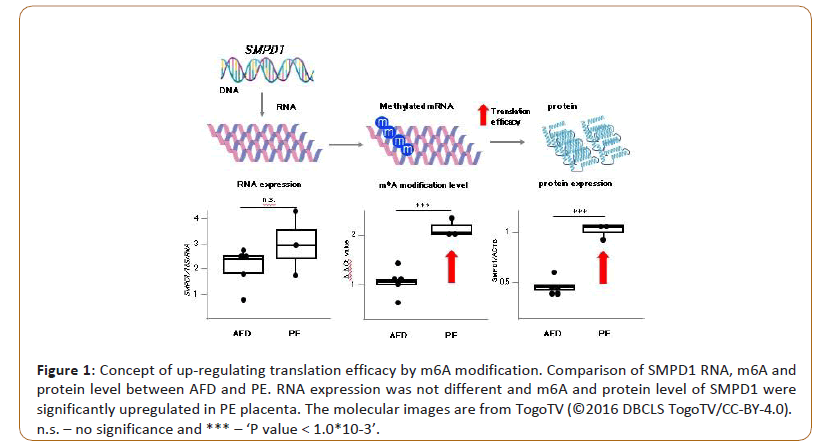
The placenta is an essential organ of the clade Eutheria (including humans), which forms a barrier between the body of the mother and the fetus, and functions to provide nutrition, gas exchange, and elimination of waste products. Placental single-cell analysis revealed that proliferation, differentiation, and regeneration are maintained even in normal placenta [11]. Therefore, cellular and molecular analyses of postpartum placentas provide useful information regarding the tissue differentiation process, which could include the long-term functions of the placenta throughout pregnancy. FTO (fat mass and obesity-associated gene) is the first GWAS-identified obesity-susceptibility gene. A cluster of single nucleotide polymorphisms (SNPs) in the first intron of FTO showed a highly significant association with the body mass index [12]. However, these SNPs were revealed to regulate the expression of distal genes IRX3 and IRX5, rather than FTO [13]. In addition, as mentioned above, FTO was identified as a demethylase of m6A [14]. Expression levels of FTO in placentas are positively correlated with birth weight [15] and prenatal fetal head circumference at 34 weeks gestation [16]. Considering the aforementioned findings, we hypothesized that m6A in placental mRNA constituted an important mechanism in post-transcriptional regulation, and was related to fetal development and pregnancy-associated diseases. Therefore, we conducted MeRIP-Seq on human placental villi obtained from mothers of infants of various birth weights and a representative pregnancy-associated disease, Preeclampsia (PE), profiled the m6A epi-transcriptome of these specimens, and investigated the relationship between fetal development or PE and placental m6A. Through MeRIP-Seq of 17 placentas derived from Appropriate For Date (AFD), Small For Date (SFD), and heavy for date, and PE with SFD, we revealed that m6A at the 5’UTR was unique in each phenotype. In concordance with the results from other types of tissue and cells, m6As are also most abundant in the vicinity of the stop codon in placental mRNAs. Although the total levels of m6A at the 5’UTR were much lower than those in the vicinity of the stop codon, we thought that the unique m6A pattern at the 5’UTR in placental mRNAs could characterize fetal growth and PE. m6A at the 5’UTR reported to have most tissue-specific m6A sites compared with other transcript regions [17]. As one of the examples is m6A modifications at the 5’-UTR mRNA could fine-tune the protein level in accordance with the pathology, we found that placenta from PE had more abundant m6A at SMPD1 (sphingomyelin phosphodiesterase 1) mRNA. Between PE and normal placentas, the expression levels of SMPD1 transcripts were similar. However, the protein level of SMPD1 was significantly higher in placentas from PE (Figure 1). We revealed that m6A modifications within two GGACH motifs at the 5’UTR of SMPD1 mRNA positively regulate SMPD1 protein expression. SMPD1 is an enzyme involved in ceramide synthesis. Ceramides are known to modulate cell signalling and drive insulin resistance, apoptosis, and fibrosis [18]. Our study suggests that there are many unknown disease-related candidate genes that m6A can regulate translation efficiency in accordance with the onset of disease without changing the amount of transcript.
Figure 1: Concept of up-regulating translation efficacy by m6A modification. Comparison of SMPD1 RNA, m6A and protein level between AFD and PE. RNA expression was not different and m6A and protein level of SMPD1 were significantly upregulated in PE placenta. The molecular images are from TogoTV (©2016 DBCLS TogoTV/CC-BY-4.0).
n.s. – no significance and *** – ‘P value < 1.0*10-3’.
In addition to our study, several studies examining m6A mRNAs of human placenta have been published. Yang et al. [3] revealed that the cytotrophoblast and syncytiotrophoblast layers of the placenta of PE with SFD had more abundant m6A in mRNAs than that of normal pregnant women. They revealed that METTL3, one of the m6A writer proteins, and hnRNPC1/C2, one of the m6A reader proteins, was significantly upregulated in the PE placenta, especially in the cytotrophoblast and syncytiotrophoblast layers, but m6A eraser FTO and ALKBH5 did not change. METTL3 knockdown in primarily isolated trophoblasts from PE placentas significantly reduced the total amount of m6A and hnRNPC1/C2 expression. On the other hand, Wang et al. conducted MeRIPseq for PE without SFD, and healthy pregnant women using placenta villi [19]. They also revealed that METTL3 and METTL14 expression were significantly upregulated in PE placental villi, and that mRNA derived from PE placental villi had much more abundant m6A. They finally detected upregulation of m6A methylation, mRNA expression, and protein expression of HSPA1A in PE placentas [20]. Jin et al. revealed that HSPA1A mRNA newly received m6A within the 5'UTR under heat shock stress response, which enabled translation initiation by a capindependent manner [21]. In Wang’s study, HSPA1A had much m6A within its coding sequence in PE placenta. Depending on where the transcript is modified, m6A might have a different post-transcriptional effect. On the other hand, Xiao et al. reported m6A function in villi of recurrent miscarriage patients [22]. They revealed that one of the m6A erasers, ALKBH5, was upregulated in recurrent miscarriage patient villi, especially in the cytotrophoblast and syncytiotrophoblast layers, and global mRNA m6A methylation levels were lower in the RM villi. They clarified that ALKBH5 inhibited trophoblast invasion by regulating CYR61 mRNA stability, which depended on the m6A methylation state in the 3’UTR of CYR61. Based on these results, human trophoblasts might be well functionally regulated and controlled by m6A. Because a recent study revealed that the placenta had the most tissue-specific m6A modification peaks [22], the placenta may also be unique in terms of epi-transcriptome. Syncytiotrophoblast dysfunction can cause PE and fetal growth restriction [23], and further investigations are needed to reveal m6A and pregnancy- associated diseases through trophoblast research.
Discussion and Conclusion
Seventy percent of mRNAs were methylated by m6A. Moreover, some researchers are trying to reveal the association between m6A site, and eQTL and SNPs; the m6A site had many eQTL. According to GWAS studies, m6A sites also had SNPs associated with HDL cholesterol levels, body mass index, and atherosclerosis. As mentioned above, we may have many important candidate genes that are missed without epi-transcriptome analysis. We have to integrate transcriptome data and post-transcriptional analysis to reveal molecular biology and pathology.
Conflicts of interest
We declare no conflicts of interest associated with this manuscript.
Acknowledgements
This mini-review was supported by Grants from the NCCHD of Japan (2020B-21).
References
- Meyer, Saletore, Zumbo, Elemento, Mason, et al. (2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149: 1635-1646.
- Desrosiers R, Friderici K, Rottman F (1974) Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proceedings of the National Academy of Sciences 71: 3971-3975.
- Yang Y, Hsu PJ, Chen YS, Yang YG (2018) Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell research 28: 616-624.
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, et al. (2015) 5â?² UTR m6A promotes cap-independent translation. Cell 163: 999-1010.
- Kislinger T, Cox B, Kannan A, Chung C, Hu P, et al. (2006) Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell 125:173-186.
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, et al. (2015) m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347: 1002-1006.
- Meng TG, Lu X, Guo L, Hou GM, Ma XS (2019) Mettl14 is required for mouse postimplantation development by facilitating epiblast maturation. The FASEB Journal 33: 1179-1187.
- Mendel M, Chen KM, Homolka D, Gos P, Pandey RR, et al (2018) Methylation of structured RNA by the m6A writer METTL16 is essential for mouse embryonic development. Molecular cell 71: 986-1000.
- Ivanova I, Much C, Di Giacomo M, Azzi C, Morgan M, et al (2017) The RNA m6A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Molecular cell 67: 1059-1067.
- Zheng, G Dahl, JA Niu, Y Fedorcsak, P Huang, et al (2013) ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49: 18-29.
- Tsang JCH, Vong JSL, Ji L, Poon LCY, Jiang, et al (2017) Integrative single-cell and cell-free plasma RNA transcriptomics elucidates placental cellular dynamics. Proc Natl Acad Sci USA 114: E7786-E7795.
- Loos, RJ Yeo, G S (2014) The bigger picture of FTO: the first GWAS-identified obesity gene. Nat Rev Endocrinol 10: 51-61.
- Claussnitzer, M Dankel, S N Kim, K H Quon, G Meuleman et al (2015) FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N Engl J Med 373: 895-907.
- Jia, G Fu, Y Zhao, X Dai, Q Zheng, et al (2011) N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7: 885-887.
- Bassols, J Prats-Puig, A Vazquez Ruiz, M Garcia Gonzalez, M M Martinez Pascual, et al (2010) Placental FTO expression relates to fetal growth. Int J Obes 34: 1365-1370.
- Barton J Mosquera, M Cleal, J K Fuller, A S Crozier (2016) Relation of FTO gene variants to fetal growth trajectories: Findings from the Southampton Women's survey. Placenta 38: 100-106.
- Zhang, H Shi, X Huang, T Zhao, X Chen, et al (2020) Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res 48: 6251-6264.
- Summers, Chaurasia, B Holland (2019) Metabolic Messengers: Ceramides. Nat Metab 1: 1051-1058.
- Gu Y, Chu X, Morgan, Lewis, D. F. Wang (2020) Upregulation of METTL3 expression and m6A RNA methylation in placental trophoblasts in preeclampsia. Placenta 103: 43-49.
- Zhou, Wan, Gao, Zhang, Jaffrey, et al (2015) Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526: 591-594.
- Li XC, Jin F, Wang BY, Yin XJ, Hong W (2019) The m6A demethylase ALKBH5 controls trophoblast invasion at the maternal-fetal interface by regulating the stability of CYR61 mRNA. Theranostics 9: 3853.
- Xiao, S Cao, S Huang, Q Xia, L Deng, et al (2019) The RNA N(6)-methyladenosine modification landscape of human fetal tissues. Nat Cell Biol 21: 651-661.
- Huppertz B (2011) Trophoblast differentiation, fetal growth restriction and preeclampsia. Pregnancy Hypertens 1: 79-86.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences